MicroRNA based switches
Discussion between Omei and I from today. Added a few puzzle links as I made the puzzles later.
Tonight I have been playing around starting from the question of what a switch would look like if based on a microRNA.
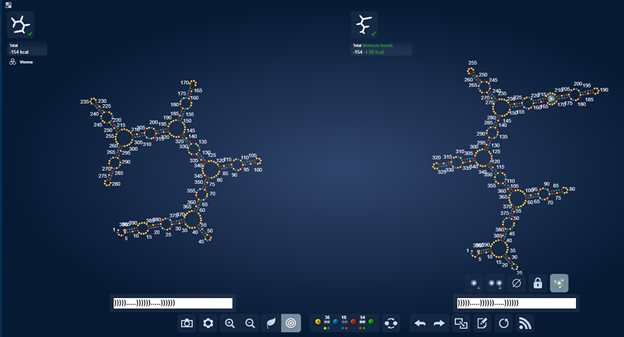
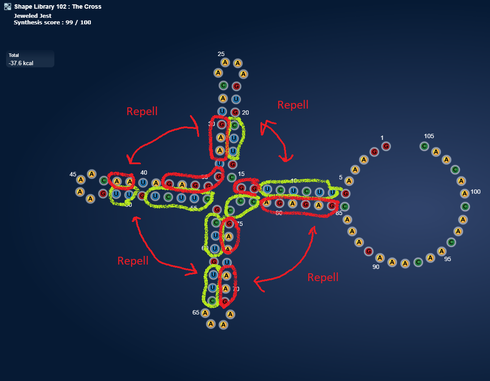
I took our old friend hsa-mir-208a and it sparked a pretty structure. I think it is perfect for creating a massive 15 state multistate puzzle.  [Correction 16 states: https://www.eternagame.org/web/puzzle/8599451/ )
[Correction 16 states: https://www.eternagame.org/web/puzzle/8599451/ )
omei [12:05 AM]
I was thinking we used a short sequence from hsa-mir-208a as an input oligo. Have I forgotten something?
eli [12:06 AM]
That is correct
omei [12:07 AM]
For the current fun, are you using just that, or the whole mir?
eli [12:07 AM]
![]()
https://www.eternagame.org/web/puzzle/8599447/
eli [12:08 AM]
Just that. I tried one of the whole unmature microRNA and it was boring.
Most of the mir’s I tried so far are boring - not sparking much structure.
Our mir was special in that it has 5 A’s. The A’s seems to help spark extra structure. [Repeat A’s and U’s seems to help with that by sparking loop structure in Vienna]
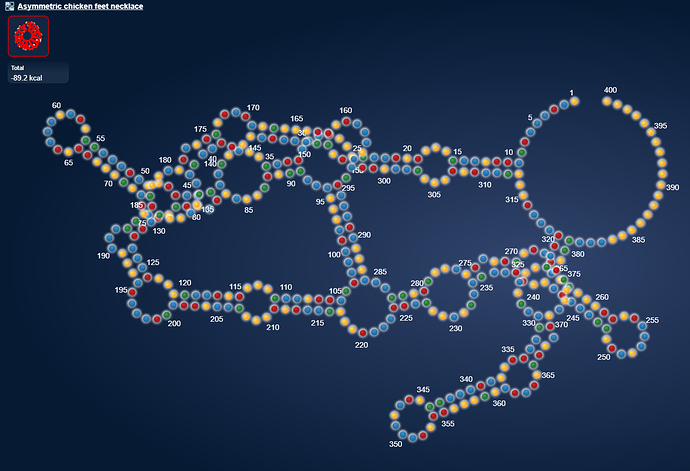
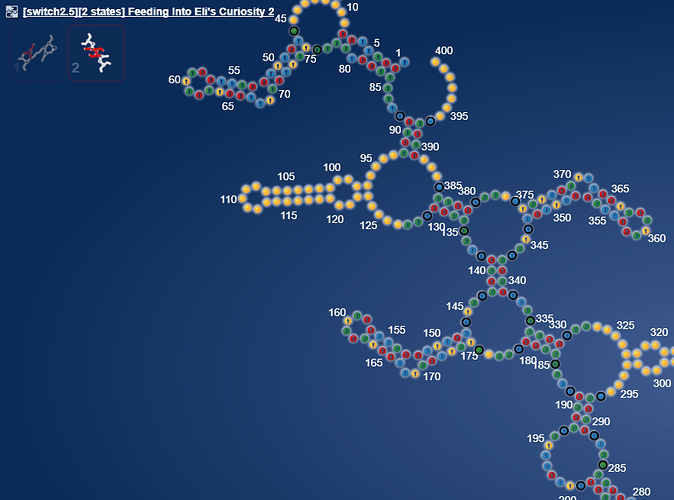
But what made me really excited now, is a whole other microRNA.
eli [12:09 AM]
https://www.eternagame.org/web/puzzle/8599478/
eli [12:10 AM]
This one is perfect for a massive switch.
I found it in a paper on alzheimers and microRNA’s that are upregulated in alzheimers.
I think it is pretty. I wonder if microRNA’s can pair with themselves.
I mean not individually.
omei [12:15 AM]
When you use the word miRNA, are you thinking about the ~22 base segment, or the ~60 base hairpin it is cut from? (the pre-miRNA.)
eli [12:16 AM]
22 base segment
omei [12:17 AM]
So our last picture is based on ~22 base segment that itself forms a hairpin?
Maybe not – I’m not sure why I thought that, looking at your picture.
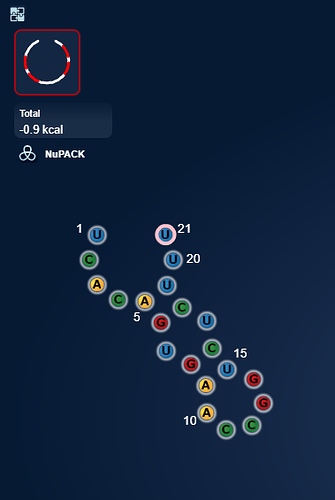
eli [12:20 AM]
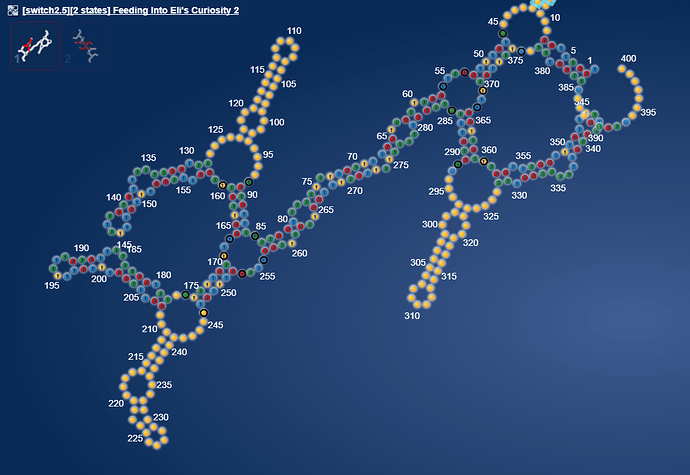
uploaded this image: Only when in NuPack
eli [12:20 AM]
But it harbours sequence that is complementary with itself if the sequence is in double.
omei [12:20 AM]
Ok, a relatively week hairpin
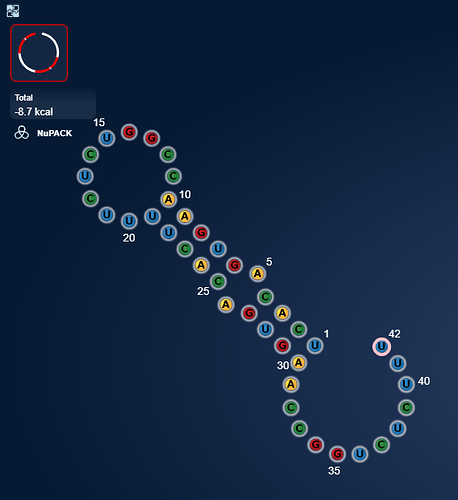
eli [12:21 AM]
uploaded this image: image.png
![]()
eli [12:21 AM]
I think my surprise stems from that I hadn’t expected anything beautiful forming from the microRNA’s. As they seem very repetitive.
omei [12:24 AM]
Repetitive in what sense?
eli [12:24 AM]
In that they are biased in sequence. Perhaps lacking one of the base letters.
Or very heavy in U’s
omei [12:25 AM]
I see.
eli [12:25 AM]
By the way U’s seems to be heavily used in full moving switches.
Example from Let-7 with no C’s:
ugagguaguugguuguauagu
omei [12:27 AM]
I suppose one “reason” for this is that they would be less likely to form dimers. (edited)
eli [12:27 AM]
Good point
I like. That could explain.
Also the sequence is cut from what is a hairpin stem and more specifically one strand of it. (edited)
And a strand in a hairpin is different in sequence from a full hairpin stem.
omei [12:29 AM]
But long single strands that form hairpins are also able to form dimers.
eli [12:29 AM]
Plus if the microRNA is too varied in sequence - it will be a good and very static stem.  Less likely to detatch.
Less likely to detatch.
omei [12:30 AM]
… which makes me wonder if pre-mRNAs have anything, like chaperone proteins, to discourage dimers.
Looks like they do require one or more proteins in oder to escape from the nucleus.
eli [12:37 AM]
Is RNA dimers kind of like palindromic RNA
omei [12:37 AM]
Exportin5 looks like it would probably distinguish against the dimer form. https://www.ncbi.nlm.nih.gov/pubmed/14730017
Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. - PubMed - NCBI
RNA. 2004 Feb;10(2):185-91. Research Support, Non-U.S. Gov’t
Not sure what you’re including in palindromic. Dimer itself means nothing more than two RNA binding to each other.
eli [12:39 AM]
Ok
Got it
omei [12:40 AM]
Any sequence that simply forms a hairpin can also easily form a dimer with another copy of itself.
eli [12:40 AM]
And since pre-miRNA are the same, they should in principle be able to bind with each other also. I think I get your point
And some miRNA’s were also downregulated in alzheimers. I may try make a puzzle of a dimirized preMRNA of one of those.
omei [12:44 AM]
It should work.
Remember in the the MS2 cooperativity puzzles, how the MS2 hairpin sequences tended to pair up against each other rather than forming a hairpin?
… in the absence of the aptamer boost.
eli [12:47 AM]
Yup.
It also seemed to matter what were in between them
In round 1 I placed stems between them. They were not happy.
In round two what worked was them being close together seperated by loop bases, and a weak stem at top.
So a loop is a very different seperator, compared to stems.
omei [12:49 AM]
In retrospect, I think the most cooperativeness might come from putting two directly adjacent. We now know the energy model incorrectly estimates the energy of adjacent stacks, but we didn’t know that then.
eli [12:52 AM]
True. My jumping jack solves (in the RNA input labs) that had a stem between the input and reporter weren’t as effective either in the RNA input labs as the solves that had the reporter and the input fairly close to each other and forming a loop between them as they turned off. (edited)
The input and reporter also there benefitted from being close together (coaxial stacked) when both were turned on.
omei [12:55 AM]
As I recall, one of the highest, if not the highest, cooperativity score came with two MS2 hairpins separated by a single base. We now know that’s close enough to get a small coaxial stacking bonus, but not nearly as much as having them adjacent.
eli [12:55 AM]
Ah, I didn’t realized that. Well noticed.
omei [12:56 AM]
Unfortunately, I don’t remember the specific design. It was from a player who was not one of the “regulars”.
eli [12:56 AM]
I think I looked more of the scores.
As in EternaScores.
omei [12:56 AM]
Right.
eli [12:56 AM]
Yup. I recall there being a new player getting a real fine score
Christopher something