@DigitalEmbrace, from top of my mind the human RefSeqGenes I have been handling so far have had a range from around 10000 to 240000 bp. +/-
There will likely be ones that have extremely long introns. I think I recall one gene that took real long time to make. Looked it up and it is Titin. It actually isn’t too bad:
In such cases, I would just up the filter level. As of now we can’t do complete scans. The pseudoknots we find are going to be used for finetuning pseudoknot finding algorithms. So as long as I get a proper amount of pseudoknots for a gene, I’m happy, even thought I know it is not a complete scan. For a short gene, I want pseudoknots enough = low filter level. For really large genes, I just want the largest pseudoknots, as they will likely also be the best targets.
I totally agree with submitting a specific gene variant of a disease that has ones interest. Thumbs up on that.
Thx for the UCSC website link. Looks complex, but potentially also very interesting. Especially gene network and the linking to outside resources.
For F8 I found an paper from 2010 saying that:
“At present, more than 1209 mutations within the F8 coding and [untranslated regions have been identified and listed in the F8 HAMSTeRS mutation database: a comprehensive international database, HAMSTeRS (The Hemophilia A Mutation, Structure, Test and Resource Site), which lists hundreds of mutations yielding the hemophilia phenotype established and maintained in the United Kingdom.”
Molecular genetics of hemophilia A: Clinical perspectives
So many different mutations of the same gene, can end up giving the same disease.
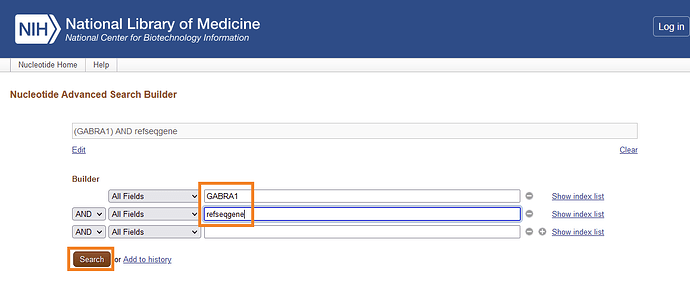
Ah, I see you meant to find the coding sequence with X01179 only. The way I would get to the mRNA via NCBI is to do this:
And then I would pick variant 1. They are all refseq’s.