I have made a system that can computationally score both elements and fragments from lab designs based on their shape data. The aim of my tool idea is to help with the growing amounts of data that is coming our way and to help us make better designs. I made the system to get the shape data displayed in the game as numbers and to help dev to mimic a player’s analysis style. I gave all nucleotides an individually numbered score after how well they did in the shape data. This analysis method can also be used together with a RNA folding algorithm.
Really cool
Being able to easily extract the quantitative bonding data from the lab results would be a big help in this regard. I tried doing some manually (just for tetra loops) once before and it was very time consuming.
http://eterna.cmu.edu/get/?puznid=165…
I mean to say, the data is there, as a JSON object if I’m not mistaken…
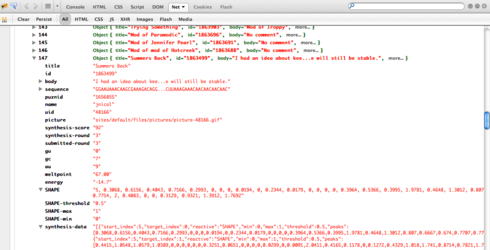
Cool. To work with the Computationally Selected aspect, we need one more datum for each lab result: a quantitative characterization of the SHAPE data. This could be as simple as a string that matches the sequence in length but contains characters (0-9, 0-9a-f, a-z, or whatever, depending on the binning/range) that indicate the degree of bonding measured for each nucleotide. If some bases were not measured, then some other character (e.g. ‘.’) can represent don’t know.
Oops: for switch data we need the quantitative SHAPE data for BOTH no-FMN and FMN-added forms. I suppose the DMS data would not hurt either, although the SHAPE data seems more crucial.
New guy here. Is there any information that might help me catch up on the program functions and objects? Hopefully I can contribute something.
Jnicol has programmed a tool based on Mat’s idea for a lab data analysis tool. Here is a link to the program:
Computationally Selected Elements
You can use the tool to help you with lab data analysis and when you are designing for lab. With it you can pick out good sequences for a particular element in a design. You can look at certain type of loops, bulges or strings.
For how to use it, see this intro.
OTHER DATABASES FOR ELEMENT SEARCH
I really want to see Rfam’s data like SHAPE data. I would love be able to search and pick out a whole element from there, like I can with Mat’s Computationally Selected Elements tool. I know Jnicol is working on an update so we can search all labs at once for a specific element.
I asked Rhiju if there were other RNA databases, where one can search for one element in many different designs. Eg. say I want a hairpin of this size or a bulge of that one. I know I can do sequence search, but I want element search, so I can use the info to cut and paste a RNA design. (FrankenRNA as an eterna player called it jokingly) If not this data is already available via Rfam or other RNA databases, it would sure be interesting having it. That would mean truly building with the building blocks of nature.
Rhiju said: There are motif databases like RNAMotif and SCOR – I don’t think they’re updated, but worth a look.
I’m kind of blown away that the players are creating these tools on their own. This is AMAZING.
We have data for some RNAs of known structure here:
and the number of natural RNAs in here is rapidly growing. Perhaps some of this data could be available in the players’ FrankenRNA-like efforts?
Also check out JandersonLee’s LabDataMiner, that is inspired by Mat’s CSE idea and Omei’s Cloud Lab Data Mining Tool.
Both tools are valuable and helpful for both designing in lab and for data analysis. Omei’s tool has its particular strength on individual lab basis. So it is great for comparison, if you are interested in only one lab. Though it can be well used for comparison on similar labs too.
Jeff’s tool is extremely helpful for digging up particular elements across our whole lab database. Its strength is comparison between large amounts of data and across lab search. It is a valuable tool for finding and reusing good elements for designing and for cross lab analysis.
Coming back to this idea nearly 10 years later. Are there any good sources for either natural and/or synthetic RNA with well defined/verified structures? Ideally I’d like multiple MB/GB of matched sequence and structure data such that I could pull out millions/billions of examples of stems and loops and possibly even pseudo-knots.
There still isn’t a ton of data for verified RNA structures, assuming you want data a step up from chemical probing. That’s a big part of what the OpenKnot project is working to address. Probably the best database is NAKB.org, which launched earlier this year.