It’s my goal to run all previously synthesized designs through several folding programs and determine the relationship, if any, between a design’s synthesis score and its performance in a given program. I’ve looked at a few designs from “The Finger” with RNAfold, and at a glance there seems to be a enough of a trend to keep me interested. If anyone wants to help out, that would speed things up quite a bit.
RNA structure prediction software is frequently used by EteRNA players to aid sequence design, but there is currently little information to support or reject its use. Though some of the programs used by players are compared in a 2004 paper by Gardner and Giegerich and ranked by CompaRNA, it is not clear that this information is relevant or useful to EteRNA players.
The data sets used in these comparisons may not be representative of the RNAs designed in EteRNA, and the “accuracy” of a program in the context of EteRNA is defined differently than in literature. In the BMC Bioinformatics paper and CompaRNA, the accuracy of programs is related to their ability to reproduce structures that have often been determined by x-ray crystallography or NMR. In EteRNA, the usefulness of a given program is based on its ability to predict designs that will receive a high “synthesis score” based on SHAPE data.
Quasispecies, this is a very important comparison that I think a lot of scientists would be interested in. We’d do it ourselves, but are short on manpower!
I’m actually wondering if you and other EteRNA players could write a scientific paper on it (I’d be happy to help edit). I think it would have a high impact on both EteRNA and the broader scientific community…
batch processing the successful designs through vienna looked possible
@ Rhiju - Sure, I’d definitely be interested in doing that.
@ Edward - Oh, yes. There’s no way I would enter all of that manually.
Update: Earlier, I ran a few of the best and worst designs for “the finger” through RNAfold and it looked like there was a correlation between synthesis score and things like ED and MFE%. I just ran all of the designs for “The Finger” that had synthesis scores and SHAPE data through RNAfold. Here are the crude results for the finger. Glancing over things it seems as if RNAfold is somewhat hit-or-miss. I’m interested to see how the other labs do.
Here’s the format of the output:
>Sequence name – synthesis score
Sequence
MFE Structure [MFE structure energy]
Crude representation of pairing probabilities [ensemble energy]
Centroid Structure [centroid structure energy, distance]
Frequency of MFE in ensemble, ensemble diversity
Not fnding the document in the “results” link.
me neither
Try it now.
Hi Quasispecies!
It is still not working. It is propably something with sharing options in google doc. I had trouble too with my own google documents, if I just clicked the option share with people who have the link. But if I choose share with all, then there were no problems.
I like your idea.
Guess I needed to update the link after changing the sharing settings. Try it one more time, and cross your fingers… the link above should work but here are new ones just in case. Please let me know if they work or not.
I’ve been able to access the files ok. Of course interpreting them is something I’ll need to spend more time on. At a quick glance, the Excel graphs look like shotgun blasts, only more random.
Thanks for taking on this project, Quasi. Let me know if I can help.
Plot of MFE Frequency in Ensemble vs Synthesis score for all labs

Plot of Ensemble Diversity vs Synthesis score for all labs (log scale)
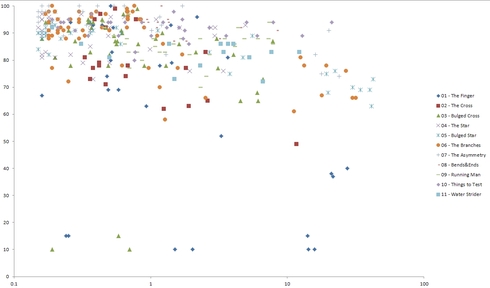
Ensemble Free Energy vs Synthesis score for all labs

The data and plots for individual labs can be found by following this link. If you want the raw data including postscripts w/pairing probabilities, I can get that to you as well. I think the next step will be to try and get at that data and explore other features of RNAfold. Once it’s all together actually do some analysis at the end. Until then, it’s probably not appropriate to pass final judgment on RNAfold in EteRNA…
Yeah, I’d love to involve other people in this. Let me know if you have any ideas or programs you’d like to try out. If you want to do stuff with RNAfold, I’m open to ideas on how to do something with the pairing probabilities data. Also, the Vienna package contains other features that no one has really looked at.
You made an earlier post on the shape of the melt curve, so you might be interested in RNAheat. RNAsubopt - a suboptimal folding program - might be interesting too.
Certainly none of these look to the naked eye like very good correlations. Posslbly because we are trying for success, and are trying for results at one end of the scale for all these numbers. Since this last lab seems to have some deliberately bad designs, maybe this will help the correlations. It looks right now to be a slight positive correlation to MFE and a negative one to ED, with no discernable trend for EFE. So the best predictor would be some combination of the MFE and ED. Maybe this shows why CompaRNA doesn’t think RNAfold is particularly good. Especially since a lot of people were using prediction software on the last 2 labs, and got pretty high values with Viennafold particularly, although neither lab was solved. It would be interesting to continue trying to solve these labs, since we have a lot of unsuccessful designs already.
UPDATE: 12/7/2011
Still working on dotplot data from past labs. All labs have been run through RNAheat, a program that generates specific heat curves, but I haven’t got around to really pouring over all of the data. A brief disclaimer - for this one I used a precompiled version of RNAheat instead of compiling it myself. Also, I’m planning to move all of the info in this thread to the wiki.
Lab 1 Specific Heat Curves Arranged by Synthesis Score
Lab 2 Specific Heat Curves Arranged by Synthesis Score
For all graphs, x axis is temperature in degrees C, y is specific heat in kcal / mol K
Does RNAheat (or meltplots based on specific heat data) show anything interesting?
In “The Finger,” the shape of the specific heat curve seems to be a fairly sensitive, but not a specific indicator of folding accuracy. All designs with synthesis scores above 90 show a similar shape. That shape also also crops up in some less-successful designs, though less frequently. The situation is less clear in “The Cross,” though there are still features that are more common in more successful designs. When compared to poor designs from the same lab, successful designs tend to show a sharper melting peak with a higher specific heat, as well as fewer peaks to the left. The meltplots tend to show sharper melting transition and higher melting point.
What is a specific heat curve telling you?
Specific heat is basically the amount of energy you need to put into a system to raise its temperature by a certain amount. The peaks in specific heat curves correspond to temperature ranges where more energy is required cause an additional increase in temperature. These peaks represent structural transitions, with the peak furthest to the right showing complete melting of the RNA. Its position is related to the melting temperature.
_ Why don’t you show me meltplots? _
According to McCaskill’s partition function paper, Specific heat curves can be used to generate melting curves. I didn’t feel up to looking up the reference that showed how to do it, and I’m lazy, so you get the raw specific heat curves. You could probably ballpark the % of bases unpaired at a given temp by taking the % of total area under the curve up to that point. Sharp, narrow peaks at high temperature mean sharp melting transitions at high temperature.
_ What does this mean? _
The trends that seem so clear in “The Finger” are considerably more muddled in more complex designs. Right now, it’s an indication that we should be thinking about what a “good” meltplot really is, how structure influences the melting behavior of an RNA, and most importantly how we as players should used (or not use) this information to design sequences.
_ Can I see the data? _
Input files.
Contain synthesized designs for all labs through “The Backwards C” in FASTA format. Labeled with name and synthesis score. (rename from .inp extension as you want):
Raw data.
All labs through “The Backwards C” as text (.out extension just for my own reference - rename as you like):
Data in excel spreadsheets.
Column 1 is temp in degrees C, column B is specific heat in kcal / mol K. The first line before an entry is the name and synthesis score. Synthesis score is the last number in the line with the name. All designs have individual Cp curves shown. Combined graphs have been done for only a few labs.
The Finger.
The Cross.
The Branches
Water Strider
Kudzu
Shape Test
Making It Up As I Go
The Backwards C
I think we need to re-evaluate what we use these programs for. Poor performance identifies a bad design fairly reliably, but mediocre to good scores don’t really seem to tell which is the better of two designs.
For example, most designs with synthesis scores of over 90% have Ensemble Diversity < 1 or MFE > 0.7. That said, designs with very high MFE% or very low ED do not necessarily fold properly… so squeezing that extra 0.02 out of ensemble diversity is probably not worth it.
The next step for RNAfold is probably to look at these two values in combination - if you have low ED and high MFE% - what is the relationship to synthesis score? Maybe using these values together is more effective than looking at either separately.
I’ve played some with RNAshapes on recent labs:
http://bibiserv.techfak.uni-bielefeld…
While I don’t have a large enough sample to be statistically significant, all of the designs that I tested in RNAshapes which had a lab value of at least 95 had a probability of folding to the desired shape of at least 0.9996 according to RNAshapes “Shapes Probabilities” analysis. However having a probability of at least 0.9996 was not a sufficient condition to guarantee a successful lab score. The winning candidate did not always have the highest predicted probability for success. Thus RNAshapes modeling might prove useful for eliminating some candidates that are likely to fail the wet labs but not to predict which are most likely to succeed.
I tested all of the synthesized candidates from the Backwards-C lab as well as a few samples from prior labs.
I’ve got data for CentroidFold with CONTRAFold and McCaskill parameters. Not sure how to use it, seeing how there are fewer simple metrics along the lines of “MFE%” and “ensemble diversity” that I could compare with synthesis score.
I can mainly think of one way to use the data - base pair distance from the predicted structure to the target structure. I haven’t really had much luck in writing anything that can take efficiently handle the output for doplots.
Just eyeballing the data, though, CentroidFold looks a lot like every other tool we’ve used in the past. It can identify garbage fairly reliably, but give it two sequences with synthesis scores > 80 and the program doesn’t seem to be able to predict which does better. Once again, this is just eyeballing things.





