I have noticed in aptamer designs, that certain sequences in the the aptamer have a habit of pairing up with certain sequences in the design. It is actually an old problem from earlier labs too, just here it was sequences in the the hookarea that paired up with sequences in the design. The sequence I’m refering to is the twice occouring aptamer sequence with red twin nucleotides.
Notice the red twin nucleotides. Those red nucleotides have a naturally longing to pair up with any green twin nucleotides they can find nearby.
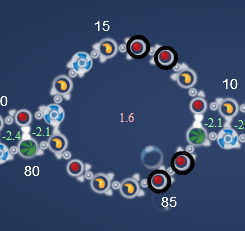
Here is the Eterna ensemble design 3 (sparse 5) (87%) from the FMN single binding site II lab, where this problem occour. The same happens for the other robots and in our designs too:
This is the structure the design should have:

Here is the estimate of how it actually folded up:

Notice how the red twin pair in the top of the aptamer paired up with the green twin pair in one of the strings.

A double sameturning GC-pairs can even cause trouble normally. There is a strategy already taking care of that.
I would like a strategy that gives an extra penalty if there is green twin nucleotides in line on a string. They won’t alway pair up, just because they are there, if the rest of the design is stable, but they always pose a risk.
Give –1 pr. each green twin nucleotides present in the design, also if the green twins are placed as a continuation between two elements ( like in an adjacent multiloop). The more green twins there are, the higher posibility that one of them pair up with the red twins in the aptamer.
Also give -0.5 if red and green twins are in close proximity, like if there is max 10 nucleotides between them. This design below shows that the red twins will find green twins even far away from home. But I think the risk of green and red twins pairing up, is higher if they are relatively close to each other.
I think this is why repetitative pattern actually is bad and why sameturning double pairs are badly tolerated, even though longer strings are more forgiving. And it also gives a perfect explanation to why two sameturning double GC-pairs seemed to pose more problems than just one in the old labs. Then the one pair could not just pair up with the red twins in the hookarea, but also the one half of a double GC-pair could get tempted to pair up with the half of the other double sameturning GC-pair.
It is also bad if twin nucleotides are sort of across each other like in the picture below, but I don’t know how to formulate a penalty for that.