When I was looking through the lab results for the aptamers with the adjacent multiloop (FMN Apatamer with single binding site I and FMN Aptamer with Single Binding Site II) I noticed a pattern that seem to cause the multiloop to slide. I took a look on old labs with this adjacent type of multiloops (multiloops with no bases between the basepairs), and this pattern always causes a slide.
Here is JerryP’s design (91% ) from the FMN ASB II
Here is how the estimate folds up.
This lead me to think that maybe lines of three nucleotides of the same color, in the multiloop were a problem in itself.
Either the presence of 3 green nucleotides in line touching the multiloop or 3 red nucleotides in line. And the combination of both those.
While I was working on the numbers on the strategy, I luckily thumbed through some papers I had printed from the forum way back. And I saw that Chris Cunningham had been on to this pattern long before me. Here is the pattern he mentions in the post [What did you learn from: Lab 103 round 1](http://getsatisfaction.com/eternagame/topics/what_did_you_learn_from_lab_103_round_1#reply_4496814
)
Chris had already noticed that the three CCC caused the multiloop to slide. As he said:
”What I’ve Learned is to avoid too many of the same base in a row, especially near complicated features – near those features (bulges or pentagons or whatever), the strand of duplicates can slide and mess up my intended shape.”
As I went through the labs with adjacent multiloops, sometimes it is a line of 3 red going through the adjacent multiloop, that causes a slide or break, sometimes it is a line of 3 green. Sometimes the combination of these two causes the trouble. But not always. Sometimes these patterns were even working quite well in The cross and The one bulged cross lab.
The pattern below, first becomes a almost certain pattern for trouble in the two aptamer labs with an adjacent multiloop. Mainly because the designs are more pressured, as all the strings are not as long as those in The cross lab and The one bulged cross lab. The adjacent multiloop is more stable in those earlier labs, while the slide happens easier in the newer aptamer labs. I’m impressed Chris cought this pattern, before it became more universal. If you look closely this pattern is present too in the picture Chris took.
Picture from GenghisJones puzzle, The revolution of the mobile archer (91%) from FMN ASB II lab.
Follow how the marked nucletides pair up.
Having two lines of green and reds almost opposite each other, is a bit too tempting for the RNA, as the green and red loves to pair up. This pattern however were succesful sometimes in the early Cross lab and One bulged cross lab, as things were not so pressured. Short strings are more energeticly pressured than long ones. Long strings can tolerate more bad pattern and repetition. The more energetic tensions there is in a design – caused by structure like shorter strings - the smaller a push is necessary to make things slide or break.
The cross & One bulge cross = long strings in connection to multiloop = The multiloop is less pattern sensitive
FMN ASB I & II = short strings in connection to multiloop = The multiloop is more pattern sensitive
From this pattern I found in relation to adjacent multiloop, I can conclude that it appears that longs strings not only in themselves are less sensitive to bad patterns than short strings. Also long desensitive strings have the same affect on the ML they are attatched to. They even make the ML less sensitive to bad pattterns too. Very interesting. ![]()
THE STRATEGY:
As it is a long time I have been seing Chris Cunninghim around, I will publish the strategy and name it in his honor.
I would like a strategy for, if there is lines of red or green touching the multiloop. This will matter only for Multiloops with adjacent stacks of 4 basepairs if it has a loop bigger than a 1-1 loop in the range of 4 basepairs or less away from the adjacent multiloop. That will include the aptamer FMN ASB I & II labs and exclude the old cross and bulged cross labs.
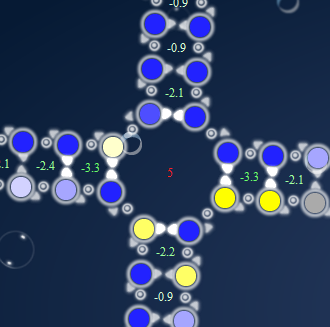
Give -1 for adjacent multiloop if such a design has both a line of 3 green and a line of 3 red touching the multiloop. Doesn’t matter which string they appear in or what side.
Give – 1 for each time a line of 3 green appears above a line of 3 reds like in this picture below. No matter what string it appears in.
While I’m at it, I will ban lines of Blue and Yellow that follows the same pattern as 3 red and 3 green in lines, touching the multiloop. I don’t remember seing it in adjacent multiloops, but I rather not see it at all, as I’m sure it will cause trouble.
Give -2 pr each of those. If there is more than 3 blue or yellow bases in line after each other, penalize with -1 pr. extra.
If there is more than 3 green bases in line, touching an adjacent multiloop, penalize with -1 pr. extra nucleotide of same color. Same with lines of yellow and blue.