I decided to try playing phylo again, as I found out that it isn’t dead like I thought and that it has an advenced mode, locked behind a screen nobody looks at (so if you played it before you might want to have a look).
While playing it, it dawned on me just how similiar phylo is to (at least my perception of) lab design. I remember toying with an idea of a script that finds the best kernel attractor of 2 oligos / oligo + MS2, and failing at a conceptual level to understand how to represent what is better than what. However, this is pretty much what phylo is, except that instead of muching R(AG) with Y(UC), we need to much the same colors, which isn’t really a problem, as the game would function just fine with anti-colored pairs instead of same-colored ones, I am pretty sure.
Ofc it would have to be changed to allow adding bases to the attractors (and to the non oligo targets), but once again, it isn’t really a conceptual problem about the game, as it could fit perfectly there, even to the style.
So what do you think? I am still a lab noob so I don’t know how much it would be needed in practice, but it sounds like it will at the very least have a niche in which it would be useful.
This is a really interesting idea. The Eterna developers of course know the developers of Phylo – we’re all RNA fans! – so they might even go for something like a ‘guest puzzle’.
I remember at some point that Phylo allowed for other scientists to put up alignment puzzles on their platform – is that a route to getting eterna data visualized on their interface? If so, is display of nucleotides A/C/G/U instead of protein sequence all we need, or is that available?
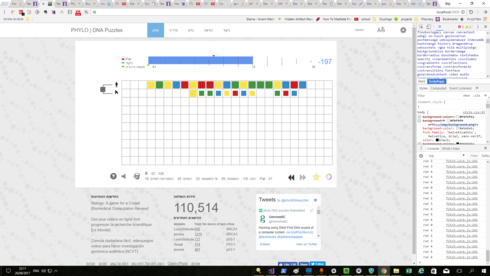
The existing 4 colors are indeed the DNA colors, and I was able to change the stylesheet to make them look like they do in eterna pretty easily.This might be enough for simple visualisation- I don’t know how much flexibility does their sequence submitor allow though, so maybe we won’t be able to do with the existing one. Also, the scoring would be very, very wrong. Will have to get to expert mode and look more at all the options to really be able to say.
I really like this idea too, MasterStormer.
Sometimes I wish to line up eg the MS2’s in a lab - which are regularly differently placed - something which isn’t possible in the existing interface.
Or I would like to line up designs after similarity of another sequence - eg like the UC sequence turnoff that exclusion labs typical have and that the aptamer and MS2 share for turnoff.
Basically lining up after similarity for a certain stretch of a design would be valuable for analysis.
eli and masterstormer – if you have a minute to try aligning some eterna design sequences in phylo, please post a link here!
This is a neat idea. I also think this could be a cool integrated tool (Foldit actually has an alignment tool built into their interface, fwiw). I can see a selection -> popup -> shift kind of thing. Could even be a booster, potentially. I don’t know if their technical infrastructure could be integrated or not (or if it would be worth it, ie for the amount of effort it would take), but at the very least we can certainly take some design pointers from Phylo.
What do you mean? A demo/explanation of how it would work?
A demo of a sequence binding perfectly to an MS2. Empty spaces are bulges.
In the end we will be the ones to decide all sequences bar a few int the top and the bottom, which will be MS2 and locked bases.
As for a word to the Phylo devs, heads up that the Phylo mobile app doesn’t increment points reliably for all users. I’ve deleted the app & reinstalled, tried old and new users, tried it on the desktop as well, with points failing to register most of the time, particularly with my old user on mobile, but I could not find a clear pattern. Hence, I assumed the project was no longer accepting data.
I looked at their tweeter, and I saw something form about a month ago. As for the score, it works like that:
You have a tree of ancestors. for every 2 children (which are also direct brothers / cousins / whatever), the game counts matches, mismatches, etc. and uses them to get a score for the “brotherhood”. Then the game creates an ancestor, to use for later*. Then the ruonds repeat, but now a single lane isn’t a “brother”, but the ancestors are. because of that the score looks weird- the lanes aren’t being matched, but the groups of them are.
*The game creates an ancestor, afaik, based on the best fitting one for the next rounds and also matches the bases of the 2 children it used (you won’t get a U in spot5 if in his children A and G were used.)
I do like the idea of being able to display oligo + MS2 or stems depending of what you wanted to display using simple boxes/bars to represent them.
https://getsatisfaction.com/eternagame/topics/ui-view-boxes
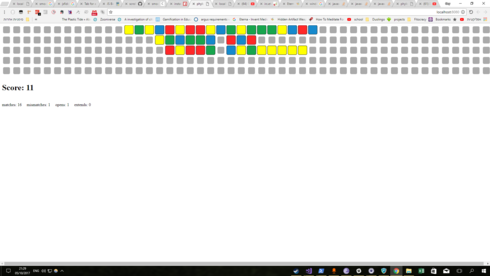
I decided to create a prototype from scratch, because I fear it would be too hard to change the phylo code to support adding new bases, and it seems that the repo isn’t maintaned. Here is what I got:
I will upload it alongside an explanation to heroku on Sunday.
Currently you can add, remove and change bases.
Sorry @mat, forgot to read your thread :X I will probably get to it tomorrow.
https://eterna-phylo.firebaseapp.com/
I will create a google docs guide tomorrow. TLDR:
- An empty spot which is agains a base is a buldge
- Right click to change the color of a base / delete it / create a new one
- The RNA piece at the top is an MS2 hairpin