I’ve explored upstream G downstream C a bit for you, here’s the picture:
The energies (excluding the tetra) are -3.1; -4.4; -4.2; did’t fold; -3.5; -2.1; -2.5; -3.5; -1.1; -2.5, the strongest being with another G-C in the end in the same direction.
Notice the one in the middle didn’t fold out. The two triplets on the right can repeat the last loop ad infinitum. An interesting side note is that i’ve put this sequence into RNAshapes and RNAfold and they predict an entirely different bulging (to this one and to each other). Here’s the sequence if you want to look for yourself:
AGGGUGGGGUUGGGUGGUGUUGGAUGGCUGGGUGGGUGGGUGACUGUCUGCCUGGCUGUCUGAUGUUGUCUGGUUUUGUUUAAAAAAA
Edit: here’s some A-U starters, first two upstream U, the rest upstream A:
Energies: -2.5; -2.5; -2.5; -2.9; -2.9; -2.5; -1.6; -2.5; -3.5; -3.5; and that could turn into infinite -2.5s too.
The two on the left are the only ones that formed with upstream U, with the second being capable of infinite looping. If you switch the G-C pair in the last one on the right, the looping stops there unless you switch some G-Us further on too.
One thing i can think of is that alternating G-U pairs have positive energy instead of negative and your RNA will want to get out of that ASAP, in effect the pairs individually attract each other, but their middles probably keep colliding and pushing apart, resulting in a bulge that is neither a connection nor a loop really.
Next picture, Gs and Us on the same side of the loop, Us upstream:

Energies: -1.2; -1.1; -3.5; -3.1; -3.5; didn’t fold; -1.6; didn’t fold
The two on the right were the only ones that formed with A-U, both needed an alternate G-U on the other end compared to the loop itself, same goes for the one that formed up on the right. This might explain why the other two on the right didn’t form up, they had A-U on the other end with no G. The only real puzzles are the two in the middle, the ones with all G-Us the same way, the single G-C doing what paramodic explained in the above post.
Even if all the above phenomena could be traced back to the alternate G-Us repulsing each other, this last one would not really be explained by that. All of these loops have negative total energies meaning that unlike regular loops which must be “forced” into shape by surrounding stacks these loops will in fact sustain themselves, and not only that they strengthen the surrounding structures too, letting you use weaker pairs around them and still sustain the shape. The real question is whether they behave the same in nature as say a 2-2 Adenine only “forced” loop or do they look and taste completely different?
And here’s a bit more:
Energies: -3.5; -1.1
The thing to notice here is that the bulge goes towards the alternating G-U regardless of the G-C present this way, to eliminate the positive loop energy that would result from that. With some G-C and A-U pairs you can force out those pairs (loop energies +0.3 and +1.3) but they will want to get away from each other no matter what.
These loops can take quite the strain too:
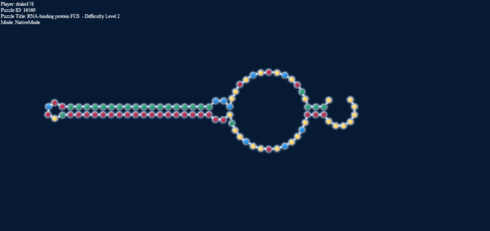
Here’s another interesting thing. The two pictures below, one in target mode with a 2-2 loop and one in natural mode without it have exactly the same energy.
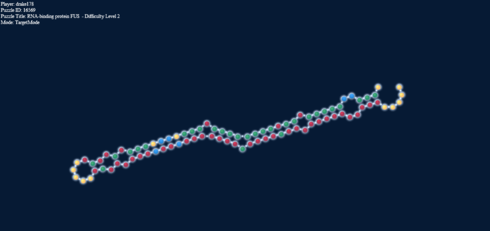
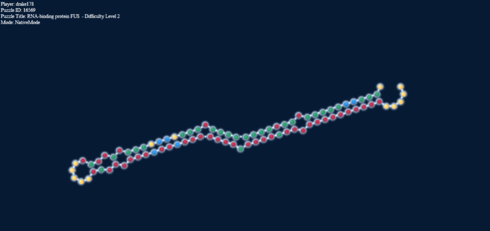
Loop energy = Paired energy = -4.1
That is, the connections formed net the same energy as the loop would. I know what you’re going to say, that would likely be the case for anything. Well, it isn’t. Here:
Loop Energy = +2; Paired energy = -5.1
But to add more to this, here’s something else: on the right we have a loop, and in the left stack, forced into target mode the identical base pairs:
Loop energy = -2.2; Paired energy = +0.1 (!)
I’ve checked a few others and they all seem to line up to the same rule, so the real question is why those G-U loops provide a greater net negative than the sum of their parts. I assume the secret lies in knowing how exactly the formed pairs relate to the bases forming right next to them. From my experience downstream double G is a stretcher for example, link a C to the first and the second will stabilize any split in the stack starting with the second G. And obviously once you’re in the middle of a strand upstream really is only downstream looked from the other side, so everything will work both ways: