I analyzed FMN MS2 Ribsowitch Round 1 and 2 labs for
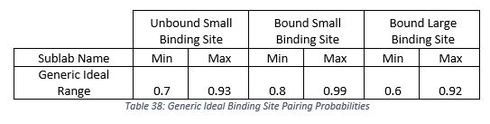
Exclusion NG 1 (EXNG1), Exclusion NG 2 (EXNG2), Exclusion NG 3 (EXNG3), Same State NG 1 (SSNG1), Same State NG 2 (SSNG2), and Same State NG 3 (SSNG3) by running all the designs through DPAT and plotting the data for all the predicted FMN binding site stacks for the small and large loop side. I plotted the minimum predicted pairing probably for each stack predicted by Vienna 2 with Nando’ FMN Hack by the EteRNA score using DPAT3. I then ordered all the plots by stack length from short too long. I then compared all the plots to find generic ideal ranges for similar target structures. I found that there are ideal FMN binding site stacks lengths and pairing minimum pairing probabilities. The best performing binding site stacks had a long static small loop binding site and a 3 or 5 base pair long large loop binding site stack in the bound state depending on how the small loops closes. If the small loop forms and actual loop it should be 5 base pairs long and if the small loop has both ends of the sequence in it then it needs a length of 3. There should not be any binding site stacks forming in the unbound state for either small loop configuration. The ideal pairing probabilities are 70% to 93% for small loop binding site unbound, 80% to 99% for small loop binding site bound and 60% to 92% for the large loop binding site bound.
Here is the paper I wrote for this. It is available as a PDF here https://dl.dropboxusercontent.com/u/87351147/Ideal%20FMN%20MS2%20Riboswitches%20FMN%20Binding%20Site…
And here is the paper without Appendix A. If you want Appendix A it is in the PDF
Also Here is the raw data this is all derived from https://dl.dropboxusercontent.com/u/87351147/Round%201%20and%20Round%202%20NG%20labs%20Raw%20DPAT3%2…
Table of Contents
I. Summary. 1
II. Analysis. 1
III. Appendix A… 32
I. Summary
I analyzed FMN MS2 Ribsowitch Round 1 and 2 labs for Exclusion NG 1 (EXNG1), Exclusion NG 2 (EXNG2), Exclusion NG 3 (EXNG3), Same State NG 1 (SSNG1), Same State NG 2 (SSNG2), and Same State NG 3 (SSNG3) by running all the designs through DPAT and plotting the data for all the predicted FMN binding site stacks for the small and large loop side. I plotted the minimum predicted pairing probably for each stack predicted by Vienna 2 with Nando’ FMN Hack by the EteRNA score using DPAT3. I then ordered all the plots by stack length from short too long. I then compared all the plots to find generic ideal ranges for similar target structures. I found that there are ideal FMN binding site stacks lengths and pairing minimum pairing probabilities. The best performing binding site stacks had a long static small loop binding site and a 3 or 5 base pair long large loop binding site stack in the bound state depending on how the small loops closes. If the small loop forms and actual loop it should be 5 base pairs long and if the small loop has both ends of the sequence in it then it needs a length of 3. There should not be any binding site stacks forming in the unbound state for either small loop configuration. The ideal pairing probabilities are 70% to 93% for small loop binding site unbound, 80% to 99% for small loop binding site bound and 60% to 92% for the large loop binding site bound.
II. Analysis
I started the analysis by running all synthesized design sequences from Exclusion NG 1 (EXNG1), Exclusion NG 2 (EXNG2), Exclusion NG 3 (EXNG3), Same State NG 1 (SSNG1), Same State NG 2 (SSNG2) and Same State NG 3 (SSNG3) for Rounds 1 and 2 through my analysis program call DPAT3. I then searched through the DAPT3 data file for each sublab and searched for all the stacks that are predicted to form that include the 3 base pairs on either side of the FMN binding site in both the bound and unbound states. For example EXNG1’s large loop side binding site stack constraint on EteRNA is stacks 9:44, 10:43, and 11:42 so any stack that contains at least those 3 base was evaluated (Figure 1: Exclusion NG 1 Large Loop Binding Site). Some stacks in the sequence of lengths do not have any designs so they are skipped.
I then plotted those stack by the EteRNA score and evaluated the plots. The list of stacks for each sublab and binding site stack using modified DPAT3 base pair notation is found in Table 1 through Table 6 bellow.
Looking at the tables for all the sublabs it can be observed that the stacks at each binding site start at a length of 3, due to the binding site base pair constraint in the UI. They then increases in size by 1 and occasionally there will not be any designs for one of the stacks and it will skip to the next nucleotide stack. An example of this is Same State NG 3 Large Loop Bound State last 2 stacks in the table. It goes from 36:81,37:80,38:79,39:78,40:77,41:76,42:75,43:74 and jumps the 35:82 base pair and then lists 34:83,35:82,36:81,37:80,38:79,39:78,40:77,41:76,42:75,43:74 as the next stack. This only occurs in longer Large Loop stacks and only in EXNG3, SSNG1, and SSNG3. ‘
Using these tables I plotted the minimum base pair pairing probabilities for each stack by the Eterna score for each design that has that specific stack and analyzed each binding site loop separately at first. Starting with the Small Loop binding site you can find all the plots for all the sublabs in order of shortest to longest stacks and unbound then bound in Appendix A. Moving on to the analysis I started with the small loop bound state for all sublabs.
I found that the shortest 3 and 4 base pair stacks had the majority of designs scoring in what I call the 30 60 group and a few with scores from 80 to 90. The 30 60 group refers to an artifact of the scoring system that ends up assigning a score of 30 and 60 for designs that are extremely bad. There are also a lot of scores that are near that range at the same time and those are included. I use a range of 20-40 and 50-70 for the 30 60 group. However there are not many designs above 80 and none that reach 100 with the only exception being SSNG2 which has a significant number of designs that score 100 with a stack length of 3 and even 4. You can see this if you review Appendix A.
It is worth noting that EXNG1 and EXNG2 do not even have any scores above 90 from Rounds 1 and 2 of these sublabs but they have a similar pattern within the dot plots. The pattern in question is that as you progress from short to long stack length the number of 30 60 group designs diminishes in general and the 30 60 score groupings tend to get a bit more tighter. Then as the stacks start getting long you start getting more designs in the 80-100 range than in the shortest stacks. Then as you reach the longest stacks there is less and less design’s in the 30 60 group. There are still bad designs in these longer stacks but they are not the score of the very bad design artifacts but I think they are the actually working switches. This indicates that there are more bad designs than good designs associated with the shorter stacks and that the longer stacks have a trend of performing better or at least having a higher percentage of design’s in a high score range. There are often high scoring designs in the stack one nucleotide pair shorter than the longest but it seems as though the longest does better more often I think. You observe this in Table 7 through Table 12. These table have the shortest, medium and longest stacks to demonstrate the transition.
The only outlier though is SSNG2 but if you consider that a design can have a 1-1 loop and still have a pretty stable stack. I looked at the next stacks in line if there was a 1-1 loop or a 2-2 loop for a 3 nucleotide base par stack and found stack 2ndState-1:84,2:83,3:82,4:81,5:80,6:79,7:78 is the stack that would form if there was a 1-1 loop just after the 3 base pair long stack. Looking at this plot for that stack you it can be seen that there are a large number of designs in the 100 score range (Figure 20: SSNG2 stack after 1-1 loop for 3 nucleotide pair long stack). In fact it is quite similar to the non 30 60 group scores from the 80’s to the 90’s. But if you look at stack 2ndState-1:84,2:83,3:82,4:81,5:80,6:79 if there was 2-2 loop it has one score close to 100 but only 2 more above 80 with a similar about under 80 (Figure 21: SSNG2 stack after 2-2 loop for 3 nucleotide pair long stack). There are just not many designs for that predicted configuration. This data however does show that having the 1-1 loop with a very long string of nucleotides pairs performs well.
My analysis software however does not yet look for combinations of stacks so it gets a little murky past these stacks. I will address this issue more a later day once my software is modified to look into this but I want to focus on the binding sites in particular today but I felt it important to show the data for the 3 nucleotide long stack in SSNG2.
Moving on to comparing the differences in the NG 2 and the remaining sublabs some things come to light. If you look at SSNG2 which has the same binding sites as EXNG2 and compare its secondary structure to SSNG1/EXNG1 and SSNG3/EXNG3 you will find that the NG 1’s and NG 3’s have a close loop that forms a hairpin but the NG 2’s do not form a hairpin since the stack has the beginning and end of the nucleotide string and forms an open loop. Knowing that the longest small loop binding site stacks are the better performers I looked at the longest predicted stacks for all the sublabs and found that they all had the same longest predicted stack in the Exclusion and Same State version of each NG sublab. The NG 1 labs had 2ndState-18:36,19:35,20:34,21:33,22:32,23:31,24:30,25:29, NG 2 labs had 2ndState-1:84,2:83,3:82,4:81,5:80,6:79,7:78,8:77,9:76,10:75,11:74 and the NG 3 labs had 2ndState-50:68,51:67,52:66,53:65,54:64,55:63,56:62,57:61. If you plug these stacks into the EteRNA UI for the sublab I found that these stack lengths correlate to the hairpin forming a tri-loop in the NG 1 and NG 3 labs (Figure 22: NG 1 Longest Small Loop Binding Site Forming Tri-Loop and Figure 24: NG 3 Longest Small Loop Binding Site Forming Tri-Loop) and a completely closed single long stack in the NG 2 labs (Figure 23: NG 2 Longest Small Loop Binding Site Forming Single Stack).
I believe based on this and the data demonstrating that longer stacks tend to do better and have more high scoring designs that long small loop binding site stacks in the bound state are ideal for FMN MS2 riboswitches. I think that the reason the NG 2 labs can do ok with a 1-1 loop is that there is another stack immediately afterward that closes the loop, which is a very stable stack. The NG 1 and NG 3 labs, however, have the tri-loop at the end which makes it a bit harder to make it a stable structure with any loop in the middle.
Next, I looked at the unbound state of the small loop binding site and checked if any of the potential binding site stacks were predicted to appear while in the unbound state. I found that once again the longer stacks had more high scoring designs. The shorter stacks were varied in their score composition but the longer stacks were the only ones with very high scoring designs. The only one to break this rule once again is the SSNG2 sublab which followed the same 1-1 loop and then long stack configuration as the bound state I previously demonstrated. You can see this in plots of all the sublabs organized by sublab with the shortest, medium and longest stacks. The plots are listed in a table for each sublab in Table 13 through Table 18.
In summary for the small loop binding I have found that longer stacks are best in both the bound and unbound state. Preferably the stack is either a fully closed stack if the NG 2 lab or forms a tri-loop if NG 1 or 3. I believe that this points to the ideal design having the small loop binding site be a non-switching portion of the secondary structure. The small loop binding site should thus form be a static stack in an ideal design.
After going over the small loop binding site I then looked at the large loop binding site in both the bound and unbound state and found that there were ideal stack lengths as well for that part of the binding site. I found that nucleotide pair stack length of 4 to 5 looks like the best I think. There are high scoring designs in the shorter stacks but there are more bad designs associated with the shorter stacks than good designs (there are a lot of designs in the 30 60 group). I think that most often lengths of 5 look best but 4 is ok but has a lot of 30 60 scores. The only sublabs that do not follow this is EXNG1 and SSNG2. SSNG2 has a lot of high scoring designs in the 3 base pair long stack. There are only a few high scores in the 5 base pair long stack. The 5 base pair length stacks does have scores only up to 90 though and this brings up one of the things I have observed. I have noticed that there appear to be groups for the high scores such as the 30 60 group and I’m not sure why.
What I mean is that there are a lot of plots that have designs that only go up to about 80 and then there are stacks that go up to about 90. Then there are the 100’s that are kinda rare. The 80 groups and the 90 groups seam to behave in the same way as the 30 60 groups in that there seems to be defined score ranges for those stacks in tight groupings. The only difference is that there is not minimum just a maximum which would be a score of 80 and about 90 respectively. There are sometimes outliers but not more than a few. This is used to pick out which of the longer stacks do better and when you look at the longer stacks only one sublab looks better with more than 5 base pairs and that is the EXNG1 sublab mentioned before. See Figure 43: 80 Score Group - SSNG1 Large Loop Bound, Figure 44: 90 Score Group - SSNG1 Large Loop Bound, and Figure 45: 100 Score Group - SSNG1 Large Loop Bound for examples of the 80, 90 and 100 groupings (the red lines indicate the max score range).
Starting on the next page are tables of all the plots for large loop binding site stack base pair lengths from 3 to 6 except for EXNG1 which will have plots for 3 to 7 base pairs due to its unique nature. The table range used for this is Table 19 through Table 24.
I then looked at the unbound state for the large loop binding site. I found that there are very few high scoring designs and they are generally only in the 90’s group. There are a two 98’s but only in one stack in one sublab out of all the sublabs. Most stacks that have scores in the 90’s group have a max score of about 85, the rest of them go up to about 90-93. The 90-93 scores are only found in a couple sublabs and there are not any scores that are 100 when there are many 100’s in the sublab for all the sublabs. There are a bunch of designs that score in the 80’s group as well.
One of the things that I have observed in the unbound state of the large loop binding site is that it is somewhat predictable but not entirely. The stacks with the majority of designs in them tend to be the ones with a base pair length of 3 to 6 and from 7 on there tends to be very few designs in the stacks for all the sublabs. The unpredictable part is which stacks will have the high scores in the 90’s and 100’s. EXNG1 has 90’s group scores in the 3 and 8 base pair length stacks, EXNG2 does not have any 90’s group only 80’s groups and there is only one score greater than 80 in the 6 base pair long stack, EXNG3 also does not have any 90’s group just 80’s groups and only 2 designs above 80 in the 4 base pair long stack, SSNG1 a single 100 group in the 4 base pair stack with a high score of 98 and a couple stacks with score sup to 86 but not enough to really consider it a 90’s group, SSNG2 has one 90’s group in the 3 base pair long stack and there are a couple mid 80’s scores but not enough to consider those stacks 90’s, and SSNG3 has all 80’s groups with only a couple scores above 80 but once again not enough to consider a 90’s group. I have listed all the plots for unbound large loop binding site due to there being so much variation. These plots can be found in Table 25 to Table 30.
I believe that all this data shows that there are ideal structures within each sublab and I feel that these ideal structures can be used to predict ideal target structures for designs with similar nucleotide length and FMN binding sites. Based on all the previous data an ideal design scoring in the 100’s group would have a static small loop binding site stack that is as long as possible which forms fully in the unbound and bound state. The large loop dynamics and a dynamic large loop binding site stack that has a 5 base pair long stacks in the bound state but ideally does not form any large loop binding site stacks in the unbound state. There are however a few large loop binding site stacks that have good scores in the unbound state but not many. What is missing in this analysis is the stacks and loops further on in the designs past the binding site stacks. I currently have a process by which you can find designs that have certain stacks and then return which of the special stacks it has in its design using DPATs analysis report function but have not performed a in depth study using that. Machine Elves observed a way to determine if loops are predicted to form correctly in the dot plots that needs to be looked into further as well and coded up.
Now that we know what stacks do best looking at those stacks I found that there are pairing probability ranges that high scoring designs are in. There are also bad designs score in the good probability ranges but the bad score also extend down past the good designs lower probability range. This means that you can filter out some of the bad designs by filtering for pairing probabilities for each stack. I found that on average the range for a good score was 60% to 100% and 80% to 100% probability. This grouping once again resembles the 30 60 and the 90’s groupings. I will call it the 60%’s and the 80%’s for the lowest that they go. None of the probs actually go up to 100% but they stay around 99%.
Below are tables of all the static long small loop binding sites and the 5 base pair in bound state large loop binding sites for each sublab showing the minimum and maximum predicted minimum probability for high scores. The exceptions are SSNG2 and SSNG3 since SSNG2 has an ideal length of 3 and SSNG 3 has an ideal length of 4 with no high scores in the 5 base par long stack for the large loop binding site in the bound state for either of them. Exclusion NG 1 has both the 4 and 5 base pair length due to it not having any scores above the 90’s group and both have the same scores so I do not know which is ultimately better and there may not even be any 100 group scores possible for EXNG1. There are definitely little things that each sublab does a bit differently than the other sublab so the following info can be used to tailor any filtering to a specific sublab or similar sublab. See the next page for the table and plots. The red lines are signify the min and max probability for high scores and the yellow line signifies the lower limit for outliers in the 90’s or 100’s group. The yellow is not the best but there are good scoring designs in that range just not a enough to consider a pattern compared to the high score ranges. See Table 31 through Table 36 for the pairing probability plots and the colors lines describes above. All probabilities in the plots are percentages in decimal form.
The paring probability ranges for each sublabs binding site stacks in the bound and unbound state are given bellow in Table 37: Optimal Pairing Probabilities for Individual Sublabs.
Looking at these plots you can see that the high scores designated by the red lines all are in about the same range as the same binding state in the same bound or unbound state as the other sublabs. From this I was able to figure out a generic ideal range for the binding site stacks to use. These are given in Table 38: Generic Ideal Binding Site Pairing Probabilities.